- ✅ 09/23–09/27/2025 — Result presentation at the MICCAI Conference in Daejeon
- ✅ 08/22/2025 — Closing of test stage submission (Extended)
- ✅ 08/15/2025 — Closing of validation stage submission (Extended)
- ✅ 08/07/2025 — Opening of test stage submission
- ✅ 06/15/2025 — Opening of validation stage submission (Two submissions are allowed)
- ✅ 05/15/2025 — Publication of downstream fine-tuning and evaluation code
- ✅ 04/15/2025 — Publication of the website & pre-training codebase
- ✅ 04/01/2025 — Release of dataset
Challenge Overview
The Self-Supervised Learning for 3D Medical Imaging Challenge aims to identify the most effective self-supervised learning (SSL) strategies for 3D medical imaging by providing a standardized and transparent evaluation framework.
Progress in the field has been hampered by inconsistent research practices — including differences in pre-training datasets, model architectures, and evaluation protocols — making fair comparisons difficult. This challenge addresses these issues by offering:
- A unified pre-training dataset
- Two fixed model architectures
- Standardized fine-tuning schedules
- A common evaluation setup
By aligning these critical components, the challenge enables rigorous benchmarking and aims to drive innovation in self-supervised learning for 3D medical imaging.
Dataset Overview
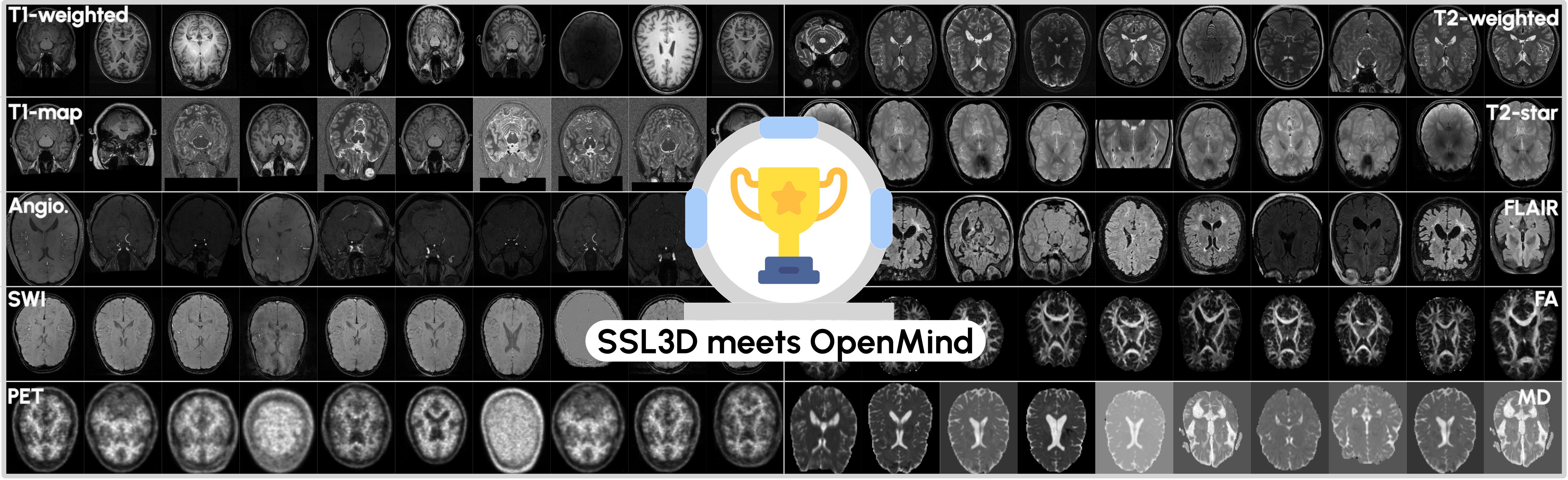
Participants will have access to the largest publicly available head & neck MRI dataset, curated from the OpenNeuro platform. This dataset:
- Combines over 800 smaller datasets, all released under CC0 or PDDL licenses
- Has been cleaned and standardized in terms of image format and metadata
- Contains 114,570 3D volumes from 34,191 patients
- Includes a wide range of MRI sequences, such as:
- T1-weighted (T1w)
- Fluid-attenuated inversion recovery (FLAIR)
- T2-weighted (T2w)
- Mean Diffusion (MD)
- Fractional anisotropy (FA)
- Additional, less common sequences
The dataset is publicly accessible via Hugging Face.
More information is available in the associated preprint: 👉 OpenMind
Task Description
Participants are invited to develop self-supervised learning (SSL) methods that enable models to learn robust and generalizable feature representations from 3D medical images. The challenge includes two predefined network architectures:
- ResEnc-L — A state-of-the-art convolutional model for 3D medical image segmentation
- Primus-M — A ViT-inspired 3D transformer architecture that closely matches ResEnc-L in performance
We provide a codebase that allows for a easy integration of new SSL methods: nnssl. It is inspired by and downwards compatible with nnU-Net. Checkout the Getting_Started to use the OpenMind for SSL pre-training.
The challenge is structured into two independent tracks, one for each architecture. Participants may choose either or both tracks.
Participants will submit only the pre-trained model weights. Fine-tuning and evaluation will be conducted internally by the challenge organizers on hidden head & neck MRI segmentation and classification tasks.
Details on fine-tuning schedules, data preprocessing, and evaluation protocols will be shared soon — see the Timeline for updates.
An initial validation stage will allow participants to test their SSL models on publicly available datasets, providing feedback and enabling iteration before the final evaluation.
Evaluation and Ranking
The official challenge ranking is computed separately for each track (ResEncL and Primus-M) as follows:
Segmentation
For each segmentation dataset, we compute the Dice score and Normalized Surface Dice (NSD) at a 1 mm tolerance threshold, following the implementation in the nnU-Net repository. These metrics are calculated per case and per class.
By default, nnU-Net assigns NaN to a metric if both the ground truth and the prediction are empty (i.e., no foreground is present). In such cases, we replace NaN with a perfect score of 1.
Next, for each class and metric, we average over all cases. For a dataset with two foreground classes, this yields four averaged values (Dice and NSD for each class). We then compute the mean of Dice and NSD across classes, resulting in two metrics per team for the dataset.
Each of these two metrics is ranked across all teams, and the average of these two ranks defines the dataset rank for the team.
Classification
For each classification dataset (all of which are binary), we compute AUROC and Balanced Accuracy for each team. These two metrics are ranked across all teams, and their average gives the dataset rank for that team.
Final Ranking
The final team ranking is obtained by averaging the dataset ranks across all datasets. Currently, the challenge includes:
- For Phase 1 (Validation): 2 segmentation datasets and 1 classification dataset (all public)
- For Phase 2 (Final): 4 segmentation datasets and 3 classification datasets (all private)
Note: This weighting scheme gives more importance to segmentation tasks, deviating from the originally proposed equal weighting between tracks.
Challenge Rules
- No additional data beyond the provided OpenMind dataset may be used for pre-training.
- Participants must create an account to submit.
- Participants may only use the PrimusM or ResEncL architectures for the corresponding tracks. Test the compatibility with the provided fine-tuning code and instructions.
- Only one submission per stage and per architecture is allowed per team. Multiple submissions from the same institution (e.g., the same university or research center) may be investigated.
- Final submissions must include a method description and a link to a public code repository to allow reproduction of the pre-training process.
- Members of the organizing institute (DKFZ) and institutes that provided downstream data are allowed to participate, but they are not eligible for awards.
- The following public tools are allowed to use: TotalSegmentator, HD-Bet, and FreeSurfer. If you want to use other tools, please ask in the challenge forum.
- Reproducible data curation and selection is allowed.
Rule violations may result in disqualification from awards and exclusion from the leaderboard.
Awards
The top 3 teams in each track will be awarded prize money. The current total prize pool is €2000. Each track will receive 50% of the amount, distributed as follows: 1st place – 50%, 2nd place – 30%, and 3rd place – 20%.
In addition, members of the top 3 teams from both tracks will be invited to co-author the resulting challenge publication.
Organizing Team
Constantin Ulrich* [1,4,5], Tassilo Wald* [1,2,3], Yannick Kirchhoff [1,2,13], Marcel Knopp [7,2], Robin Peretzke [1,4], Maximilian Fischer [1,4,12], Partha Ghosh [1], Fabian Isensee [1,3], Adam Hilbert [14], Paul Naser [9,10], Lars Wessel [11], Martha Foltyn-Dumitru [8], Gianluca Brugnara [1,8], Jochen B. Fiebach [15], Jan Oliver Neumann [9], Laila König [11], Philipp Vollmuth [1,8], Klaus Maier-Hein [1,2,3,4,5,6]
Contact Email: constantin.ulrich@dkfz-heidelberg.de
- German Cancer Research Center (DKFZ) Heidelberg, Division of Medical Image Computing, Heidelberg, Germany
- Faculty of Mathematics and Computer Science, University of Heidelberg, Germany
- Helmholtz Imaging, DKFZ, Germany
- Medical Faculty Heidelberg, University of Heidelberg, Germany
- National Center for Tumor Diseases (NCT), NCT Heidelberg, A partnership between DKFZ and University Medical Center Heidelberg
- Pattern Analysis and Learning Group, Department of Radiation Oncology
- German Cancer Research Center (DKFZ) Heidelberg, Division of Intelligent Medical Systems, Heidelberg, Germany
- Division for Computational Radiology & Clinical AI (CCIBonn.ai), Department of Neuroradiology, University Hospital Bonn, Germany
- Department of Neurosurgery, University Hospital Heidelberg, Germany
- AI Health Innovation Cluster, German Cancer Research Center (DKFZ), Heidelberg, Germany
- Department of Radiation Oncology, University Hospital Heidelberg, Heidelberg, Germany
- German Cancer Consortium (DKTK), partner site Heidelberg
- HIDSS4Health - Helmholtz Information and Data Science School for Health, Karlsruhe/Heidelberg, Germany
- Charité Lab for Artificial Intelligence in Medicine (CLAIM), Charité – Universitätsmedizin Berlin
- Center for Stroke Research Berlin , Charité – Universitätsmedizin Berlin
*equal contribution
Sponsors
The award money is generously provided by our partners. We sincerely thank them for their support in making this challenge possible.